|
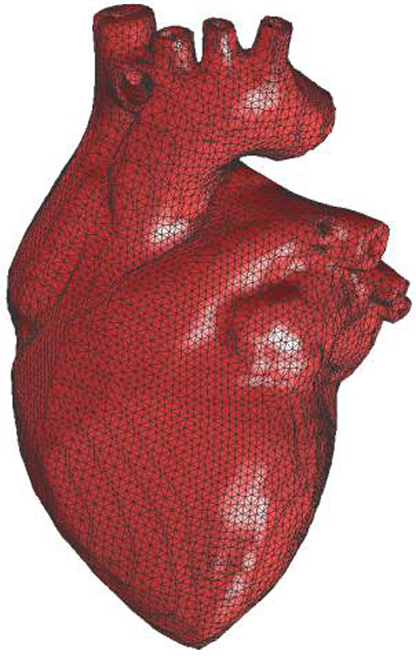
A tetrahedral finite element mesh of a human heart model.
A simple triangular surface mesh of the heart is converted
into volumetric finite element tetrahedral mesh using our mesh generation method,
now part of our software tool LBIE
Additional
images for heart and
brain modeling with tetrahedral and hexahedral meshes
Link to Movie: Heart (Movie1
),(Movie2), Brain (Movie)
|
|