|

|
Description
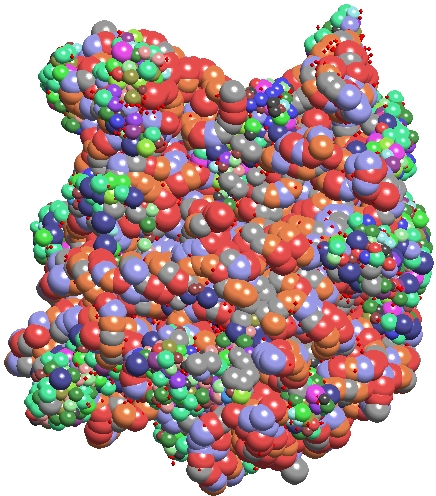
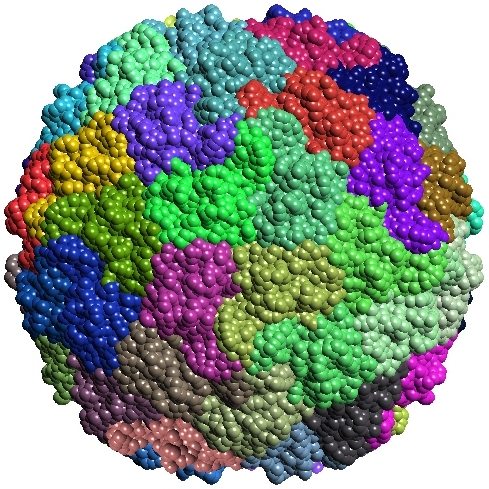
TexMol is a molecular visualization and computation package. It allows for quick and high quality visualization of Proteins and RNA, and reads in the PDB format.
- Open source software for rendering large molecule data sets.
- Written in C++, with a QT front end. The same code should work on multiple platforms, including Windows and Linux.
- Reads molecular structures in PDB and PQR formats.
- Produces high quality images using High end graphics cards functionalities.
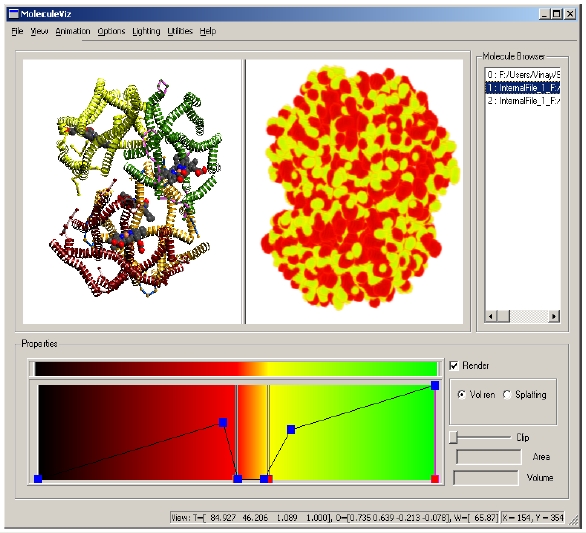
- Volume rendering and calculation of electron density, hydrophobicity functions.
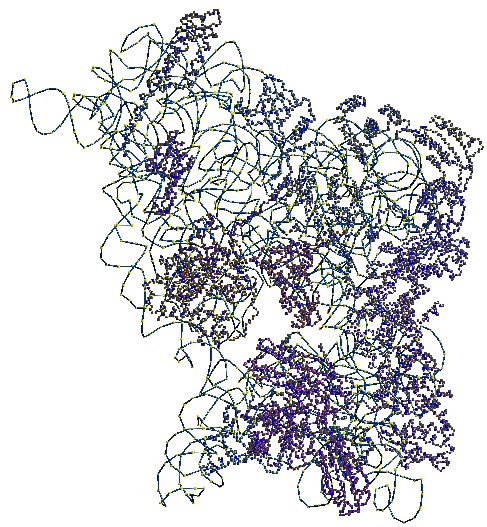
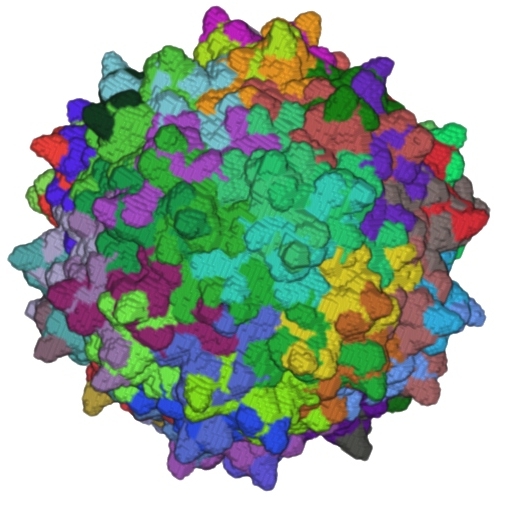
- Wireframe and smooth shaded isosurfaces.
- Computes metrics including curvatures, surface areas and volumes.
- Provides programmers with a simple and yet powerful hierarchical data structure of the molecules, with various torsion angles calculated.
- Multiple views and multiple data set rendering.
TexMol provides a user interface to a number of CVC software packages including F2Dock (protein docking), MolSurf (molecular surfaces), MolEnergy (molecular energetics), and Pockets/Tunnel Extraction. TexMol utilizes the DPG data structure to efficiently process large molecules.
|
References
C. Bajaj, P. Djeu, V. Siddavanahalli, A. Thane
TexMol: Interactive Visual Exploration of Large Flexible Multi-component Molecular Complexes
Proc. of the Annual IEEE Visualization Conference, October 2004, Austin, Texas, IEEE Computer Society Press, pp. 243-250. (pdf)
|
Download
Source packages are delivered on demand upon explicit request.
|
Software Usage
See the TexMol User Guide for documentation.
|
|
|





