|
Apart from just the shapes, we are also interested in volumetric functions,
whose contours define a better estimate of shape, and can be used for animation,
docking and other calculations. Some important fields we are interested in are
electron density and electrostatic potential.
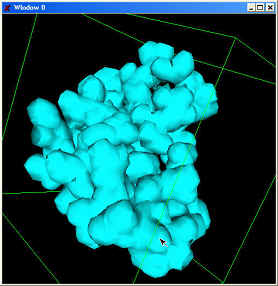
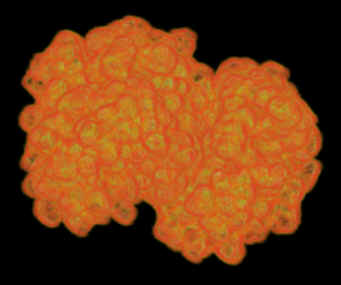
We have developed an electron density field estimation program which inputs
either PDB or PQR data and calculates a function of electron density. Gaussian
kernels are approximated around each atom, the magnitude and extent being
determined by the atom type. APBS, a package from Nathan Baker is used to obtain
the electrostatic potential calculation. One of the functions used to evaluate
this field is the sum of bonded and non bonded energies, calculated from summing
up the contributions of all atoms and their bond energies.
- Electrostatic density evaluation package which uses a Gaussian kernel to
approximate the field
 |
 |
 |
| Electron density |
Laplacian of electron density |
Electrostatic gradient magnitude |
Baker, N.A., Sept, D., Joseph, S., Holst, M.J., and McCammon, J.A.
Electrostatics of nanosystems: Application to microtubules and the
ribosome.
Proc. Natl. Acad. Sci. 98, 2001, 10037-10041.
Davis, M., and McCammon, J.
Electrostatics in biomolecular structure and dynamics.
Chem. Rev. 90, 1990, 509-521.
Honig, B., and Nicholls, A.
Classical electrostatics in biology and chemistry.
Science 268, 1144-1149 (1995).
Connolly, M.L.
Analytical molecular surface calculation.
Journal of Applied Crystallography 16, 1983, 548-558.
Max, N.L.
Computer representation of molecular surfaces.
IEEE Computer Graphics and Applications 3, 5 August 1983, 21-29.
Lee, B., and Rachards, F.M
The interpretation of protein structures: Estimation of static accessibility.
J. Mol. Biol 55, 1971, 379-400.
|